Association Testing in Related Individuals
Family data is correlated that could lead to inflation in test statistics if not accounted for. Many genetic studies contain related individuals. Family-based studies were developed to avoid bias due to population structure; Biological family members are genetically matched.
Most behavioral and environmental factors do not alter DNA, so SNP associations are unlikely to be confounded EXCEPT confounding by ancestry.
Population Stratification Bias
- For case control studies, population structure means bad matching; cases and controls come from different genetic populations.
- For cohort/cross-sectional studies with quantitative phenotypes, if the phenotype and genotype distributions both differ by population then population stratification bias can occur.
- If a population consists of a mixture of subpopulations and it is not accounted for in the analysis, false evidence for association may result.
- Spurious association only occurs when there is a difference in BOTH genotype and phenotype distribution with subpopulations.
Designs for Family-Based Studies
- Early-onset phenotypes:
- Case-parent trios (where child is effected) - Transmission disequilibrium test (TDT)
- Late-onset phenotypes:
- Discordant siblings - sub-TDT/conditional logistic regression
- General designs
- Nuclear or extended families - Family-based association test (FBAT)
We'll mostly focus on case-parent trios and TDT tests.
Case-Parent Trios
In this design we collect samples of trios; Two parents and one affected child (affection status of parents are not considered). The idea is under Mendels laws, heterozygous parents transmit each allele with equal probability (1/2) regardless of population structure. If there is preferential transmission of a specific allele from parents to affected offspring it indicates that allele is associated with the disease.
Transmission Disequilibrium Test (TDT)
Tests whether a particular marker allele is transmitted to affected offspring more frequently than expected. Non-transmitted alleles act as matched controls.
H0: No association or no linkage
Ha: Variant is associated with disease/trait (and is not due to population structure)
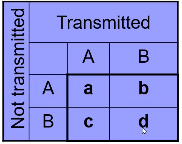
Notation:
- a = number of AA parents
- b + c = number of AB parents
- b = Number of AB parents transmitting B to affected child
- c = Number of AB parents transmitting A to affected child
- d = number of BB parents
Under the null we would expect: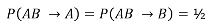
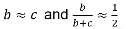
The test statistic follows a McNemar test: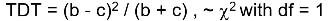
We observe this is only a function of the heterozyzgous parents, homozygous parents provide no information.
Discordant Siblings
- TDT unit of analysis is affected child + parents.
- Discordant siblings - unit of analysis is sibships with at least one affected and one unaffected sibling
- Analysis possibilities:
- Cochram-Maentel-Haenszel test
- Conditional Logistic Regression
- Like TDT these tests are conditioning on the genotypes in the family
- TDT: conditioning on the parent's alleles
- Sibs: conditioning on the observed genotypes in the sibship
General Family Structures
- Family Based Association Test (FBAT) and other similar tests (requires specialized software)
- Determine the expected genotypes of the affected individuals conditional on the family structure
- Use this to create a score test comparing observed and expected genotypes
- Same idea as parent-offspring trios or case-control sibships: conditioning on the observed genotypes
Unconditional Tests
- TDT, sTDT, FBAT and other family based association tests are conditional tests
- These tests are immune to population structure bias but often ignore information that can be used to assess association when population structure is not a concern
- Homozygous parents
- Sibships where the case and control have the same genotype
- Modern analyses can account for population structure using ancestry information available with genome-wide data
- Unconditional analyses account for the family correlation and make use of all the observed data
- These analyses tend to be more powerful than the conditional family-based tests
Review: For unrelated (independent) subjects we can use standard linear or logistic regression. Including related individuals in the analysis violates the assumption of independence, causing inflation in the test statistics.
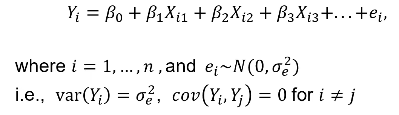
Where Xi1 is the genotype of the ith person and beta1 is the fixed affect of the SNP. Xi2 and Xi3 are covariates.
- For quantitative traits use Linear mixed effects models are frequently used for corrected observations for an individual or related individuals.
- Pedigree-specific methods use a variance component that is dependent on the kinship matrix for individuals.
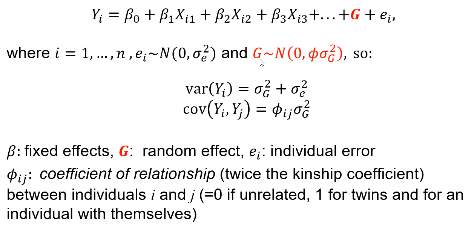
We can see the mixed effects model adds a G variable to account for genetic variance component.
For association analysis of quantitative traits, if X1 is the SNP then the test H0: Beta1 = 0 is a test of association just as for the linear model for unrelated subjects. The regression estimate beta is exactly the same as in simple linear regression.
In either case G is not tested for significance. If we want to test for heritability we estimate variance of G.
Common software: GCTA, SOLAR, R GMMAT package, Genesis R/Bioconductor package.
Dichotomous Traits
- Logistic mixed effects can be used to account for relationships
- Fit using penalized quasi-likelihood method
- Requires iterative matrix inversion - can be very slow
- Alternative: Logistic model with Generalized Estimating Equations (GEE)
- Accounts for correlated observations in a general way
- Produces a "robust" variance estimate to account for correlation
- Tends to result in inflated Type-I error
- Low frequency SNPs primary probelm
- Extent of inflation varies
In this course we focus on quantitative traits to keep things simple.

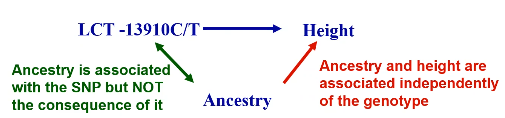
No Comments