Association Testing in Related Individuals
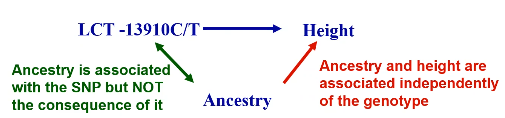
Family data is correlated that could lead to inflation in test statistics if not accounted for. Many genetic studies contain related individuals. Family-based studies were developed to avoid bias due to population structure; Biological family members are genetically matched.
Most behavioral and environmental factors do not alter DNA, so SNP associations are unlikely to be confounded EXCEPT confounding by ancestry.
Population Stratification Bias
- For case control studies, population structure means bad matching; cases and controls come from different genetic populations.
- For cohort/cross-sectional studies with quantitative phenotypes, if the phenotype and genotype distributions both differ by population then population stratification bias can occur.
- If a population consists of a mixture of subpopulations and it is not accounted for in the analysis, false evidence for association may result.
- Spurious association only occurs when there is a difference in BOTH genotype and phenotype distribution with subpopulations.
Designs for Family-Based Studies
- Early-onset phenotypes:
- Case-parent trios (where child is effected) - Transmission disequilibrium test (TDT)
- Late-onset phenotypes:
- Discordant siblings - sub-TDT/conditional logistic regression
- General designs
- Nuclear or extended families - Family-based association test (FBAT)
Case-Parent Trios
In this design we collect samples of trios; Two parents and one affected child (affection status of parents are not considered). The idea is under Mendels laws, heterozygous parents transmit each allele with equal probability (1/2) regardless of population structure. If there is preferential transmission of a specific allele from parents to affected offspring it indicates that allele is associated with the disease.
Transmission Disequilibrium Test (TDT)
Tests whether a particular marker allele is transmitted to affected offspring more frequently than expected. Non-transmitted alleles act as matched controls.
H0: No association or no linkage
Ha: Variant is associated with disease/trait (and is not due to population structure)
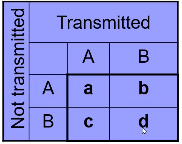
Notation:
- a = number of AA parents
- b + c = number of AB parents
- b = Number of AB parents transmitting B to affected child
- c = Number of AB parents transmitting C to affected child
- d = number of BB parents
Under the null we would expect: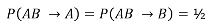
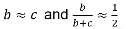
The test statistic follows a McNemar test: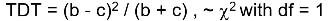
We observe this is only a function of the heterozyzgous parents, homozygous parents provide no information.