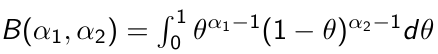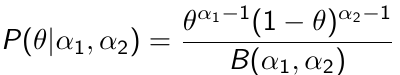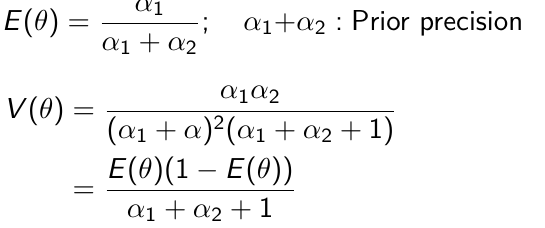Introduction to Bayesian Modeling
In the frequentist approach, estimated probabilities are viewed as one of an infinite sequence of possible data of the same experiment Bayesian analysis is based on expressing uncertainty about unknown quantities as formal probability distributions.
Bayes Problem: Given the number of times in which an unknown event has happened and failed - Requires the chance that the probability of its happening in a single trial lies somewhere between any two degrees of probability that can be named.
Bayesian statistics used probability as the only way to describe uncertainty in both data and parameter. Everything that is not known for certain are modeled with distributions and treated as random variables.
| Exposed |
Unexposed |
||
| Diseased |
a |
b |
m1 |
| Not Diseased |
c |
d |
m0 |
| n1 |
n0 |
n |
Recall the odds ratio (OR) is the probability of having an outcome (disease) compared to the probability of not having the outcome: ad / bc
Bayesian Statistics takes the point of view that OR and RR are uncertain unobservable quantities.
Prior probability: Model/probability distribution using existing data/knowledge
Posterior probability: Updated model with new and prior data
Bayes theorem allows us to learn from experience and turn a prior distribution into a posterior distribution.
Bayes Theorem
P(H) = Probability of H (Prior Density)
P(H | data) =( P(data | H)*P(H) ) / P(Data) = Posterior probability of H
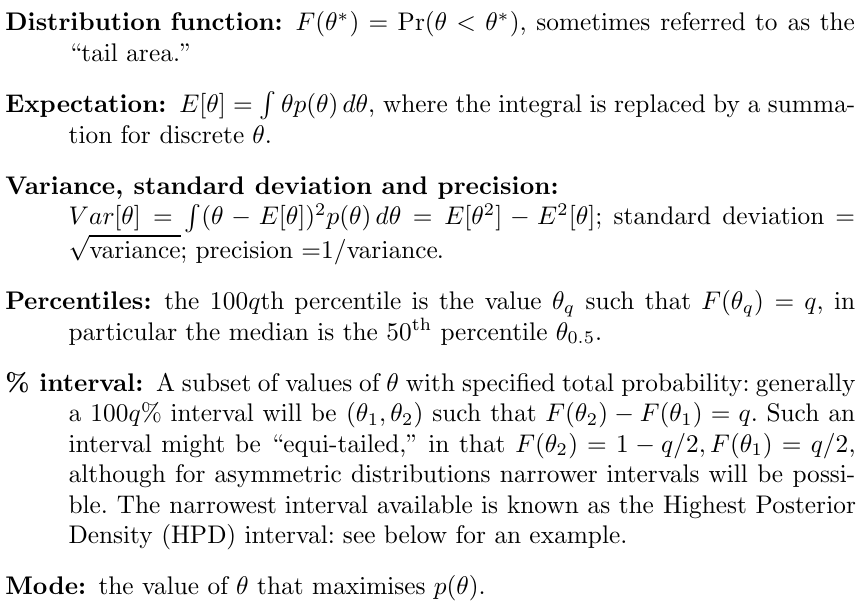
Consider a generic probability distribution p(𝜃) for a single parameter 𝜃. The probability distributions are defined as:
Markov Chain Monte Carlo (MCMC) Simulation
There are several ways to calculate the properties of probability distributions for unknown parameters. We will be focusing on using simulation of the known random variables and estimate the property of interest.
Note that there are entire classes on algorithms used for simulation, we will not go into great depth on the underlying theory.
Monte Carlo algorithms are a broad class of computational algorithms that rely on repeated random sampling to obtain numerical results.
Suppose we have a random variable X which has a probability distribution p(x) and we have an algorithm for generating a large number of independent trials x(1), x(2), ... x(T) . Then:
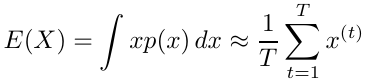
By the law of large numbers the approximation becomes exact as T approaches infinity.
Additionally, if I(l < X < u) is an indicator function where the value is 1 when X lies between l and u and 0 otherwise, the probability can be estimated with Monte Carlo integration by taking the sample average for each realization x(t)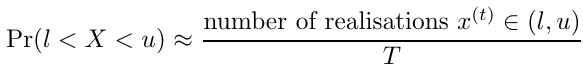
In R:
# Step 1: Generate samples
x <- rbinom(1000, 8, 0.5)
# Represent histogram
hist(x, main = "")
# Step 2: Estimate P(X<=2) as
# Proportion of samples <= 2
sum(x <= 2)/1000
## 0.142JAGS
JAGS is functionally for the MCMC. It is not a procedural language like R; it can be called from R. You can write complex model code in any order using a very limited syntax
The below code does the same thing using JAGS (in R):
library(rjags)
### model is defined as a string
model11.bug <- "model {
Y ~ dbin(0.5, 8)
P2 <- step(2 - Y)
}"
writeLines(model11.bug, "model11.txt")
# Now run the Gibbs sampling for 1000 iterations
### (Step 1) Compile BUGS model
M11 <- jags.model("model11.txt", n.chains = 1, n.adapt = 1000)
### (Step 2) Generate 1000 samples and discard
mcmc_11 <- update(M11, n.iter = 1000)
### (Step 3) Generate 10000 samples and retain for
### inference
test_11 <- coda.samples(M11, variable.names = c("P2", "Y"), n.iter = 10000)JAGS uses directed graphical models for internal representation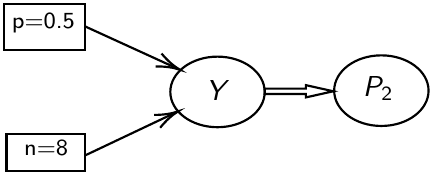
- Nodes represent variables
- Directed arrows represent conditional probability distributions
- Double arrows are mathematical functions
The joint probability distribution can be derived using Markov properties of marginal and conditional independence (next section).
Beta Distribution
Beta is a family of probability distributions with support between 0 and 1, it is the typical choice for working with probability parameters. The density function depends on two hyper-parameters 𝛼1 and 𝛼2.
Functions rbeta(), dbeta(), qbeta(), pbeta() available in R for working with Beta distributions. It's also implemented in JAGS as function dbeta( , )
The prior hyper-parameters determine the expected Y events.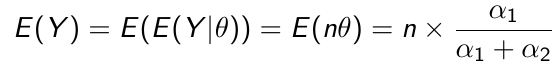
𝛼1 + 𝛼2 = equivalent sample size
Gibbs Sampling
Gibbs Sampling is MCMC algorithm that reduces high dimensional simulation to lower dimensional simulations. Used in BUGS (Bayesian Inference Using Gibbs Sampling) and JAGS as the core algorithm for sampling from posterior distributions.