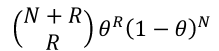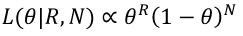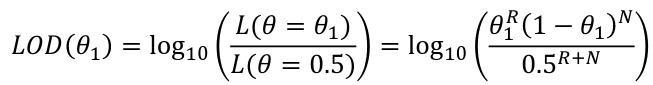Linkage Analysis
The primary goal of linkage analysis is to determine the location (chromosome and region) of genes influencing a specific trait. We accomplish this by looking for evidence of co-inheritance of the trait with other genes or markers whose locations are known, and locating genes close to one another.
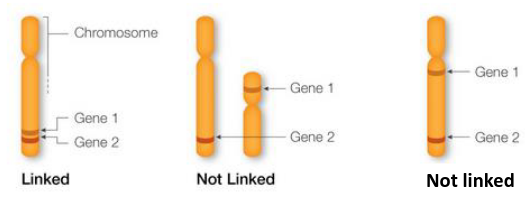
- Genes are markers that sit close together on a chromosome are called "linked" and are likely to be inherited together.
- Genes on separate chromosomes are never linked
- Genes that are far away from each other on a chromosome are likely to be separated during homologous recombination and are considered not linked
Recall Mendel's 2nd Law: The Principle of Independent Assortment - Alleles of a gene pair assort independently of other gene pairs. The segregation of one pair of alleles in no way alters the segregation of another pair of alleles EXCEPT when the genes are linked on a chromosome.
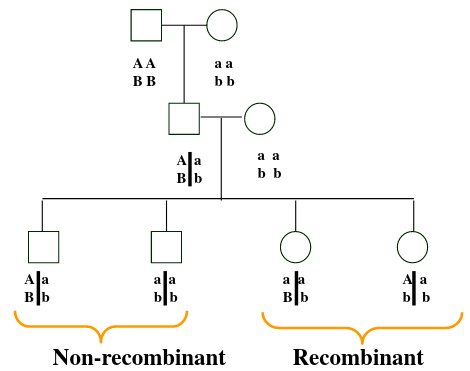
Recombinant don't inherit the same chromosomes as their parents, while non-recombinant have chromosomes inherited by their parents. Proportion of recombinants gives an indication of the distance along the chromosome between the two loci.
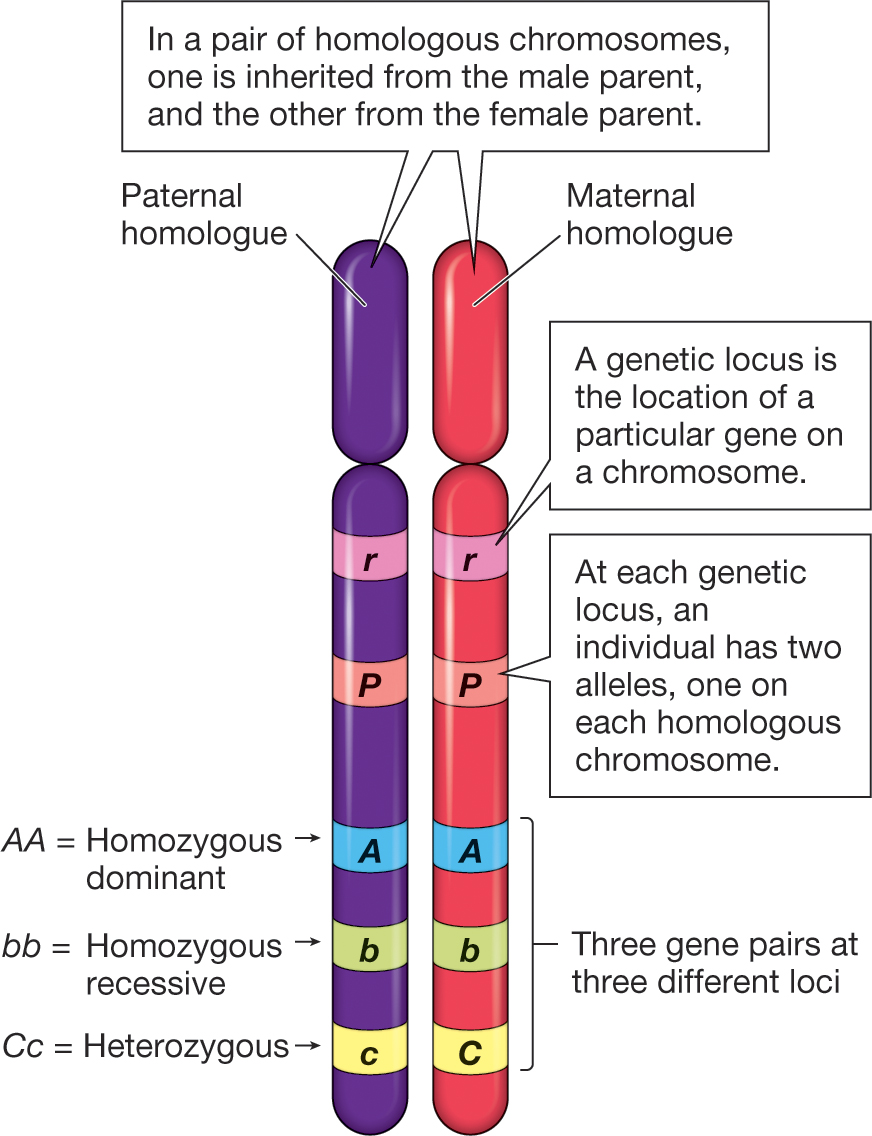
A haploid genotype (haplotype) is a combination of alleles at multiple loci that are transmitted together on the same chromosome. It contains 1) The alleles present at each locus (multi-locus genotype) and 2) Which alleles are on the same chromosome.
Recombination
The recombination fraction () is the probability of recombination (i.e. the probability of an odd number of crossovers) between two loci.
- The closer two loci are located on a chromosome the less likely they are to recombine
- The further aprart they are, the more likely they are to recombine
- 𝜃 = 0 if:
- The genetic marker is the polymorphism causing the disease
- The marker is so close to the disease mutation that recombination can never occur
- 𝜃 = .5 if:
- There is a 50% chance that alleles at the two loci are inherited together. This happens when the two loci are
- Very far apart on a chromosome
- Located on two different chromosome
- There is a 50% chance that alleles at the two loci are inherited together. This happens when the two loci are
When loci are on different chromosomes, we observe recombination 50% of the time.
The line inbetween the allele means they are on the same chromosome.
Linkage Analysis
Linkage analysis estimates the genetic distance between genetic markers or between genetic markers and a trait locus with recombination. This allows us to know where a trait locus is in the genome when we know 1) the genetic distance between a disease locus and a marker AND 2) the location of the marker on the genome.
To perform a linkage analysis, estimate the recombination fraction between the disease locus and a marker locus and test:
H0: 𝜃 = 1/2 -> not linked i.e segregating independently
H1: 𝜃 < 1/2 -> linked, i.e. close together on the same chromosome
The most common test of linkage is Logarithm of the Odds (LOD):
LOD Score = log base 10 of the likelihood ratio = log10(L(𝜃=𝜃1)/L(𝜃=.5))
This is a transformation of the usual likelihood ratio test. The probability of observing R recombinants and N non-recombinants, where the recombinantion fraction is 𝜃, is as follows:
So the likelihood is:
We can ignore the constant because we willl be working with a ratio of likelihoods and the constant cancels.
When we know the exact number of recombinants and non-recombinants:
The MLE of 𝜃 is 𝜃_hat = R/(R+N) which is the value of 𝜃 that maximizes the LOD score and likelihood function.